Some changes to your DNA are known to increase your risk of developing certain types of cancer. These DNA changes are usually found in all tissues in the body and can be passed along to your children. Using molecular tests like DNA sequencing on normal tissue (such as blood or saliva) can determine if a person has one or more of these known DNA changes that are genetic risk factors, and can therefore assess risk for developing particular cancers, decide to initiate a screening program, or help choose a targeted or proven medication.
Some examples include Lynch syndrome (an inherited predisposition to developing colorectal cancer or endometrial cancer, in addition to other types of cancers, due to changes in genes that are involved in DNA repair) and susceptibility to developing breast, ovarian, pancreatic, and prostate cancers due to changes in the BRCA1 or BRCA2 genes.
Testing to determine inherited predisposition to cancer is known as germline testing, genetic testing, or inherited disease testing.
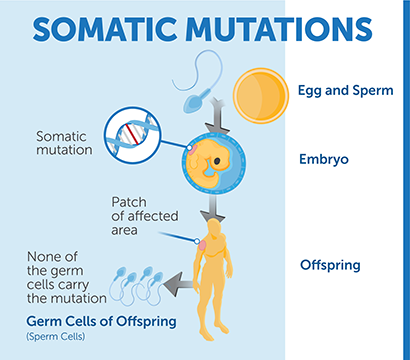
While some DNA changes are found in all tissues of a patient's body, cancerous tissues and tumors frequently have specific DNA changes, sometimes called “somatic mutations”. These changes are found in cancerous tissues and are referred to as biomarkers, they can include mutations and other genetic changes.
Determining if a patient has one or more of the known biomarkers can help inform diagnosis, directed therapy (such as specific drugs or immunotherapies), prognosis, clinical trial eligibility, and patient management. Some biomarkers may not currently have an associated therapy and other treatment options may be more appropriate.
A patient can be tested for biomarkers through molecular laboratory tests, such as DNA sequencing, performed on a sample of cancerous tissue take from a biopsy.
Some examples include testing of non-small cell lung cancers (NSCLC) to effectively guide targeted therapy and identifying chronic myeloid leukemia (CML) patients who would benefit from treatment with specific tyrosine kinase inhibitors, such as imatinib.
Testing to determine a patient's biomarker status is known as cancer biomarker testing, cancer genetic testing, molecular testing or tumor profiling.
Generally, most biomarkers are found only in the cancerous tissue and are not passed along to the person's children. However, sometimes a person will have a DNA change throughout all the tissues of their body that makes them more likely to develop cancer AND specific DNA changes that only occur in the cancerous tissue. In this case, a full “biomarker profile” for this patient would include both classic biomarkers and any genetic risk factors for developing cancer.

Currently, there are a diverse array of cancer biomarkers that are known to affect diagnosis, predict response to directed therapies (such as an FDA-approved immunotherapy), determine prognosis, inform clinical trial eligibility and impact patient management. These biomarkers can be assessed by many different techniques, including nucleic acid testing, in addition to protein-based assays or other types of analytes. Some examples of tests used to assess a person's biomarkers are below. Please note that this is not an inclusive list and each type of test is not appropriate for all types of cancer. Some of these tests can also be used to determine a person’s inherited risk of cancer.
What is DNA?
DNA is the hereditary material that encodes the information that produces the structural materials and molecules (proteins) that perform the functions necessary for cell (and organ) function. DNA is made up of individual bases (A, C, G, and T) and their order, or sequence, determines the instructions encoded by the DNA. The DNA bases pair up together: A always with T, C always with G.
What is DNA-sequencing?
DNA is "sequenced" when a molecular professional uses specialized machines and laboratory techniques to “read” the bases along the DNA strand. This information is then stored in a computer file. Depending on the unique case for each person, and the specific type of testing being performed (more information below), either small specific sections of DNA can be sequenced or all the DNA (the entire genome) can be sequenced in pieces. A specialist physician or doctoral scientist analyzes the data generated through sequencing and prepares a comprehensive results report that includes the key findings, such as detections of changes or biomarkers.
What is the difference between getting a gene sequenced and getting the whole genome sequenced?
A gene is a segment of DNA that encodes for a protein in our body that has a particular function. Humans have over 20,000 genes in our DNA, but this only represents 1% of the information in our DNA. When diagnostic laboratories use sequencing technology, depending upon the clinical question at hand, they can sequence part of a gene, a single gene, many genes, or even the entire genome (all the genes, also known as whole genome sequencing). Whole genome sequencing includes all of the parts of the DNA, not just the genes.
What is panel testing?
Molecular professionals can test single genes or all the genes, but they can also test a few genes that give them the information that is needed. The ‘somewhere in the middle’ is panel testing of two or more genes. Typically, panels are designed based upon the clinical question that is being asked. For example, a hereditary breast/ovarian cancer panel could be useful for a person with a family history of breast and/ovarian cancer. Testing for one gene (i.e. BRCA1) may be insufficient to diagnose an inherited cancer syndrome, but a panel of genes may be able to answer the question of “do I have an inherited cancer syndrome?”.
What is next generation sequencing and why is it different?
Next generation sequencing, or NGS, is an extremely powerful sequencing method that allows a clinical laboratory to sequence an incredible amount of genes at one time. Traditionally, sequencing required a separate tube and reaction for a portion of each gene and simply testing for one gene could require 8 tubes. With NGS, diagnostic clinical labs now have the ability to sequence the equivalent of millions of tubes at one time. As a result, NGS as a tool has been credited for the explosion of genetic information over the past decade.
Are there other similar tests that assess the sequence of the DNA?
Many different types of tests that assess the sequence of DNA can be used by laboratory professionals depending on the type and extent of information that needs to be obtained for the patient. Sequencing that looks for very specific changes in the DNA or more extensively at broad areas of the DNA can be performed. Sequencing of RNA, a related molecule, can also be useful in certain situations.
What is the difference between "whole genome" and "whole exome" sequencing?
When diagnostic laboratories use sequencing technology, they can sequence part of a gene, a single gene, many genes, or even the entire genome, also known as whole genome sequencing. Whole genome sequencing not only includes all of the parts of the DNA for genes (known as exons) but also includes the intervening segments of DNA between. Whole exome sequencing includes only the DNA for genes.
How long does this test take?
Multi-gene next generation sequencing panels typically take 10-14 days, with smaller panels often taking less time, and some broader panels (or whole genome sequencing) taking longer. In some cases, it is important to have a specific answer very quickly to guide therapy choices, and in these very specific cases results may be available in as little as a few hours. If you know the name of the test and the name of the performing laboratory, you may be able to find information about how long that specific test should take online in the laboratory’s test directory.
Do I need to get this test performed by my doctor or can I order it online?
If you are diagnosed with cancer or have a strong family history of cancer, this testing may be ordered by your doctor. The doctor treating your cancer makes decisions about which tests to order based on your specific situation, and your doctor may need this information to help make a correct diagnosis and/or make decisions about which treatment might be most effective.
Germline testing for inherited risk of cancer is also available online with or without physician authorization, however a more detailed diagnostic test to confirm the results may be required by your physician. Please see the Consumer Genomic Testing Versus Physician-initiated Genetic Testing section in the "FAQ About Molecular Testing" for more information.
What does FISH testing look for?
FISH (or fluorescence in situ hybridization) testing looks for chromosomal changes that are known to be common in certain types of cancer. These include chromosomal deletions, duplications, translocations (rearrangements in which chromosomes have broken apart and reassembled themselves on other chromosomes), and inversions (rearrangements in which regions of an individual chromosome have become shuffled from their normal arrangement into an opposite arrangement on the same chromosome). FISH testing can help diagnose certain types of cancers that have defining chromosomal changes, such as acute promyelocytic leukemia, as well as provide prognostic and therapeutic information for other types of cancers.
How does FISH testing work?
FISH testing uses fluorescent labels that bind to specific regions of the chromosome. Thin sections of cancer tissue from a biopsy are placed onto glass slides, submerged in these fluorescent labels, and then analyzed under a fluorescent microscope by trained molecular professionals. These professionals analyze the pattern of fluorescence observed to determine if there are any unexpected changes to the chromosomes, and if so, what the exact change is. For instance, in cancers in which parts of chromosomes are deleted or duplicated, FISH labels targeting the deleted regions will have fewer than the expected number of signals in the affected cancer cells, while FISH labels targeting duplicated regions will have more than the expected number of signals. In all cases, several hundred cells are individually counted and analyzed to determine the overall characteristics of the cancer tissue.
Is FISH testing recommended for all cancers?
FISH testing is not recommended for all cancers. It should only be used: 1) when a specific FISH label is available that can detect the chromosomal region(s) of interest and 2) when translocation(s) / gene fusion(s) or chromosomal aberrations (e.g. loss or gain of a chromosomal region) have been previously shown to have a strong diagnostic and/or prognostic association with a particular cancer. FISH can also be used to detect aneuploidy (abnormal number of chromosomes), which is also sometimes associated with specific cancers.
What is the difference between FISH testing and getting a gene sequenced?
FISH testing assesses visually by microscope the presence or absence of small sections of the chromosome. Gene sequencing, on the other hand, determines the “spelling” of an unknown sequence under study and therefore, can detect mutations (“misspellings" of the DNA sequence) as well as pinpointing the exact base-pair that the chromosome change occurred at.
How long does this test take?
In general, FISH testing is performed within a few days, but can take up to two weeks.
Do I need to get this test performed by my doctor or can I order it online?
This test is ordered only by your doctor. The doctor treating your cancer makes decisions about which tests to order based on your specific situation, and your doctor may need this information to help make a correct diagnosis and/or make decisions about which treatment might be most effective.
What does “Microsatellite Instability” mean?
Microsatellite instability occurs when specific proteins that repair mistakes in DNA, called mismatch-repair proteins, are not present or do not function properly. As a result, every time a cell divides, many, many mistakes (also known as mutations) are present in the DNA of each new cell.
Lack of functioning mismatch repair proteins have been identified in many types of cancer, and scientific studies have demonstrated that these cancers may respond to drugs that help the person’s immune system attack the cancer cells, often referred to as immunotherapies. One example of a cancer syndrome that results from not having functional mismatch proteins is Lynch syndrome, an inherited syndrome that results in higher likelihood of a person developing a variety of cancers. An oncologist might suggest that a person with colon cancer be screened for microsatellite instability and Lynch syndrome in order to decide on the best course of treatment. It is important to know that microsatellite instability is uncommon or rare in most types of cancer.
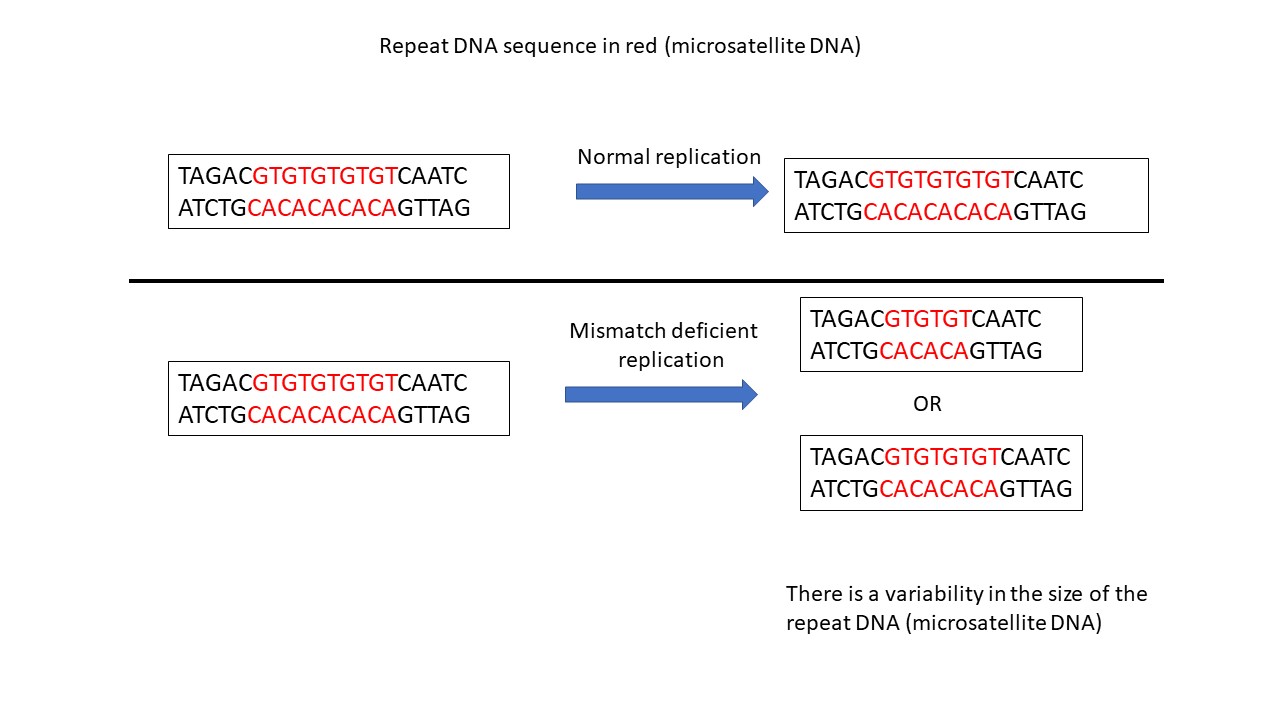
To determine if a tumor does not have functioning mismatch repair proteins, laboratory professionals look for how many mistakes are present in the DNA of a tumor sample. Instead of checking for mistakes throughout the entire genome, professionals focus on specific areas that have repetitive sections (for example, GTGTGTGTGTGTGT) and are known as “microsatellites”. It is known that mistakes frequently occur in these repetitive, microsatellite regions during normal cell division and are quickly fixed by the mismatch repair proteins so they do not cause harm and are referred to as “stable.” When there are no functional mismatch repair proteins, any mistakes that naturally occur in these areas are not fixed, resulting in errors in these regions in the new cells (illustration below). When cells have variations in the repeat length of these microsatellite regions, they are considered to have “microsatellite instability.”
How is MSI assessed?
Microsatellite instability (MSI) can be assessed by looking at the repeat lengths of microsatellite DNA in tumors compared to non-tumor tissue. Tumors are classified as MSI-high when more than 30% of the microsatellite sites are abnormal or as MSI-low when less than 30% of the microsatellite sites are abnormal. Laboratory professionals use a variety of different techniques to assess the repeat lengths of the microsatellite regions.
Is MSI testing recommended for all cancers?
Microsatellite instability is associated with cancers that are related to an inherited cancer-predisposition syndrome called Lynch syndrome. The most common cancers associated with Lynch syndrome are colon cancer and endometrial (uterine) cancer. Current guidelines are that all colon and endometrial cancers should be assessed for mismatch repair deficiency to determine if a patient should undergo further evaluation for Lynch syndrome. Assessing for Lynch syndrome is important in order to understand a person’s risk for other Lynch-syndrome associated cancers in themselves and relatives.
Beyond Lynch syndrome, an oncologist might want to evaluate a person with cancer for microsatellite instability to determine if they are eligible to receive specific immunotherapies. Thus MSI testing can be considered for any advanced or metastatic cancer where immunotherapy is a possible treatment.
What is the difference between MSI testing and getting a gene sequenced?
MSI testing evaluates specific segments of DNA that have repetitive sequences. When a cancer has defects in mismatch repair, the repetitive sections of DNA will become unstable, with random additions and subtractions of the repetitive sequence, resulting in differences in the number of repeats from cancer cell to cancer cell. MSI tests assess the lengths and variability of these repetitive regions of DNA in cancer tissue samples, often comparing the repeats against non-cancerous tissue, to determine if there is greater variability in the number of repeats that could be explained by mismatch repair deficiency. In contrast, when a gene is sequenced, the laboratory professional assesses for changes in a specific genetic sequence of the gene to determine if a mutation is present that would be predicted to result in a change or loss of function. Thus, MSI testing is evaluating the effect (phenotype) of mutations in mismatch repair genes, rather than evaluating the sequence changes in those genes.
How long does this test take?
Typically, this test takes between 7-28 days (average is 14 days). Additional time may be necessary for confirmatory or additional tests.
Do I need to get this test performed by my doctor or can I order it online?
This test is not offered online. It must be ordered by a medical practitioner, if considered appropriate. Genetic counseling may be recommended if the results suggest a defect in the mismatch repair genes that could be inherited. Additional testing of a blood sample may also be indicated. This additional test would be performed to check for mutations in relevant mismatch repair genes that may occur in the person’s germline. This information would be important to determine recurrent risk.

Learn how mutations matter for Cholangiocarcinoma patients with this fantastic video, presented by The Cholangiocarcinoma Foundation. Please visit the website below for more information.
Learn about the new, evidence-based clinical practice guideline on biomarker testing for Colorectal Cancer. This video was produced by the American Society for Clinical Pathology (ASCP), College of American Pathologists (CAP), the American Society of Clinical Oncology (ASCO), and AMP to explain what the guideline is, what is included in the guideline, and what this means for patients.
Learn about the updated and revised Molecular Testing Guidelines for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosnie Kinase Inhibitors. This video was produced by the College of American Pathologists (CAP), the International Association of the Study of Lung Cancer (IASLC), and AMP to explain what a guideline is, why this guideline was updated, what changed in the guideline, and what this means for patients.
Created: 08/2020 Last Updated: 08/2020